Our paper entitled “A Systematically Optimized Miniaturized Mesoscope (SOMM) for large-scale calcium imaging in freely moving mice” has been published in Nature Biomedical Engineering.
The recording of neuronal population activity of freely behaving animals has been typically based on cellular resolution head-mounted microscopes with a conventional microscope design. For such microscopes however, typically limitations have been posed stemming from hindrances of field of view (FOV), resolution, weight and optical aberrations: Some large-scale neuronal imaging approaches have addressed aspects of miniaturization and weight to enable free animal behavior. In other instances, head-mounted miniaturized microscopes have achieved sub-millimeter FOV at the tradeoff of low depth of field (DOF) and susceptibility to motion induced artifacts due to axial focus drift. Despite these advances in free behavior imaging, a functional microscope design containing a combination of multi-millimeter FOV, cellular resolution, tolerable weight, and robustness against axial drift in freely behaving rodents does not exist.
In this work, we present a systematically optimized miniaturized mesoscope (SOMM), a widefield, head-mounted fluorescent mesoscope based on a principled optimization approach that allows for mesoscale, cellular resolution imaging of neuroactivity while offering robustness against motion-induced artifacts. Our design enables recordings of neuronal population activity at up to 16 Hz within a FOV of 3.6 × 3.6 mm2 in the cortex of freely moving mice while featuring 4-µm resolution, a DOF of 300 µm at a weight of less than 2.5 g.
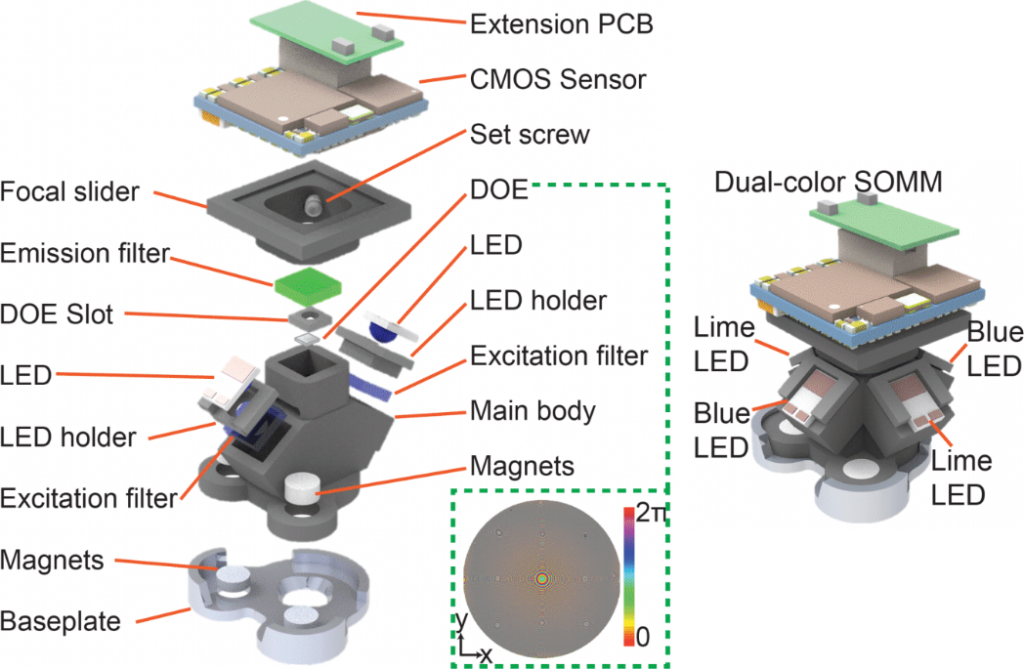
Figure 1 | SOMM Setup. A 3D decomposed view of the miniature mesoscope components in detail. The DOE phase profile wrapped into [0, π] is illustrated in the bottom right corner. The phase profile which was critical for the optical design and optimization for maintaining the FOV and resolution of a theoretical ideal lens design in practical neuronal population recordings. CMOS, complementary metal-oxide semiconductor. PCB, printed circuit board. LED, light emitting diode. DOE, diffractive optical element. Right, 3D rendering of dual-color SOMM.
The inception of SOMM has yielded an optimized head-mounted miniaturized mesoscope which incorporates a single diffractive optical element (DOE) while offering the best possible combination of FOV, minimized form factor, weight and illumination (Figure 1). We also realized the SOMM to offer dual-color imaging capability.
To achieve this, we designed and created an optimization scheme which identified the optimal tradeoff of these collective parameters as a set of fixed criteria. The optimization scheme can be broken down into distinct steps for this process. To optimize the axial form factor we had to satisfy a design governed by the angle of ray incidence onto the pixels, neuron-resolving resolution and satisfaction of the Nyquist theorem by ensuring diffraction-limited spot size is sampled by two or more camera pixels and at least two times smaller than neuron size. By assuming an ideal lens when assessing the tradeoffs for the lens we found the largest FOV which minimized form factor (Figure 2a). Although the design parameters identified amounted in a ~40% smaller axial footprint and ~50 times greater FOV, aberrations are not considered in an ideal lens model.
To properly factor aberrations for a large FOV a deconvolution algorithm with a DOE was implemented in favor of the ideal lens model. In this method the ideal lens design was approximated by identifying an optimized joint solution for the DOE phase profile (Φ) and deconvolution parameters (λ) which maximized the FOV for single-cell resolution. Furthermore, the specifications of the axial point spread function (PSF) for the DOE was optimized such that single-cell resolution would be maintained in the instance of axial motion.
Utilizing realistically simulated volumetric cortical tissue we were able to achieve the iterative optimization of these parameters through an optically informed convolutional neural network (CNN) (Figure 2b). With our forward model we tested different PSFs by convolving them with the realistically simulated models, projected the output to the axial (z) direction as a representation of a detected image. Next, we calculated the L2 loss between deconvolved images and the axial summation of brain tissue and back-propagated the result to update the weights and biases of the CNN model. Forward and back propagation were performed until the parameters of the model were optimized, therefore yielding a DOE phase profile capable of maintaining a FOV and resolution of a theoretical lens, while being suitable for neuronal population recordings. Moreover, the PSFs generated by the optimized DOE are axially elongated which has eliminated the need for 3D scanning when recording neurons in the axial direction across the 300-µm depth (Figure 2d).
We developed a custom data processing pipeline for SOMM, which incorporates our SV-Deconv algorithm and the previously published DeepWonder package, a neuronal signal extraction technique for widefield fluorescence recordings.
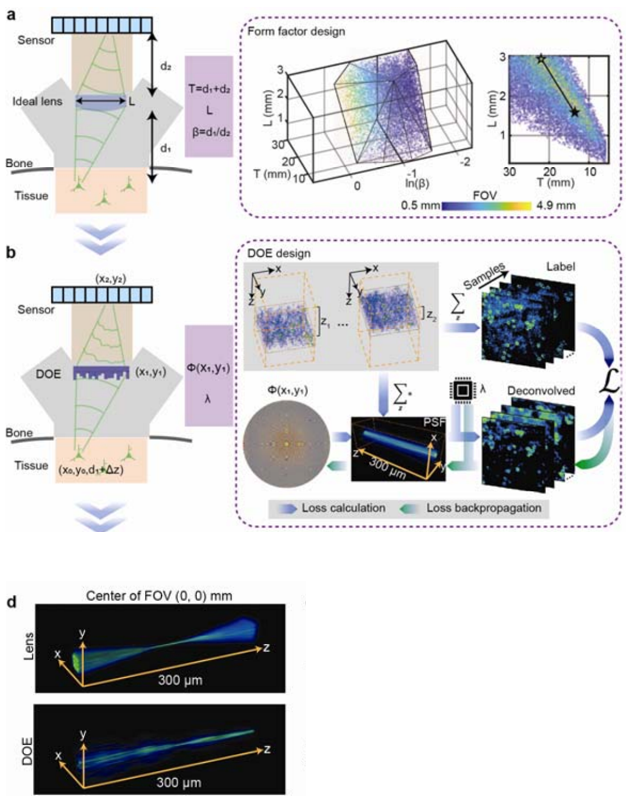
Figure 2 | Sequence of jointly and iteratively performed optimization scheme. (a) First optimization round of SOMM: A minimal form factor and maximal field-of-view (FOV) for an ideal imaging system were identified through a joint search of the parameter space while satisfying constraints for feasible angles of light incidence onto pixels, neuron-resolving resolution, and a pixel sampling ratio to fulfill the Nyquist’s limit (Methods). The optimized variables include sample-to-lens distance d1, lens-to-sensor distance d2, and imaging aperture L. To simplify the optimization, the equivalent variables T = d1 + d2, β = d1/d2 and L were optimized instead. The feasible solutions of T, β, and L and were scattered and formed a manifold where different colors represented different FOVs (from 0.5 mm to 4.9 mm). (b) Second optimization round of SOMM: The parameter space for a joint solution for diffractive optical elements (DOE) and corresponding deconvolution algorithms that approximated the ideal lens scenario in a. was searched. To achieve this, we fed simulated volumetric neuronal tissue to the DOE-based imaging system via nonparaxial propagation (forward model, blue arrows; Methods), and computed the loss between deconvolved captures and depth-averaged virtual tissue (Methods). The loss was then back-propagated to update both DOE phase φ(x,y) and parameter of the deconvolution algorithm (backward model, green arrows). The simulated volumetric samples were generated using NAOMi (Methods), and randomly shifted across a 3.6 × 3.6 mm2 FOV and 300 μm DOF to resemble large-scale imaging conditions during free behavior with axial drift. (d) 3D rendering of the point-spread function (PSF) of a plano-convex lens (top) and the optimized DOE (bottom) at the center of the FOV. The plano-convex lens had the same focal length and aperture as the optimized DOE in SOMM.
Next, ground truth verification was performed to demonstrate and statistically quantify the accuracy of the neuron positions and the neuronal temporal signals recorded by SOMM. To do so, mouse cortex was imaged using a hybrid setup that allowed to split fluorescence between a SOMM device and a standard two-photon microscope which served as a simultaneously acquired ground truth modality. We found excellent agreement between SOMM and ground truth, even when the z-plane imaged in two-photon was located 150 μm away from the center of the SOMM imaging volume, demonstrating the large depth of focus achieved by SOMM (Figure 3).
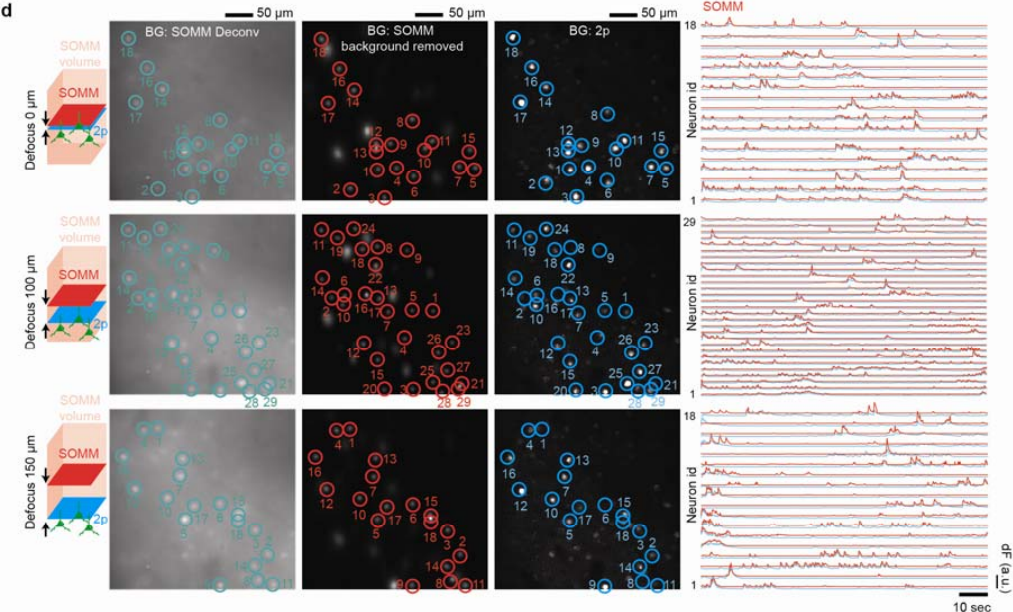
Figure 3 | Spatial and temporal evaluation of SOMM with 2p functional ground truth. Temporally interleaved 2p functional ground truth was generated by interleaving SOMM frames and planar 2p microscope frames (Supplementary Fig. 18a, Methods). First column: schematic plot showing the relative positions of 2p (blue) and SOMM (red) focal planes within the SOMM detection volume (pink). Second to forth columns: STD of deconvolved SOMM video, STD of background removed SOMM video through DeepWonder [39], and STD of 2p video, for 3 different SOMM defocus configurations (top: 0 μm; middle: 100 μm; bottom: 150 μm; see details on defocus configuration in Supplementary Fig. 18b). SOMM-extracted neuronal positions are shown as circles overlaid on the deconvolved (second column, green) and background-subtracted videos (third column, red). Paired 2p neuronal positions were analyzed by CaImAn followed by human annotation, and were overlaid in the 2p videos (forth column, blue circles). Fifth column: neuronal activity traces from SOMM (analyzed by the proposed pipeline, red) and 2p (analyzed by CaImAn, blue) corresponding to circles in left panels, as used for performance quantifications.
We then used our SOMM system to capture large-scale in vivo neuronal population activity in transgenic mice expressing layer-2/3-specific GCaMP6f calcium indicators. Recordings in freely behaving animals engaged in social interactions demonstrated high-contrast activities of thousands of neurons (Figure 4) (Video 1).
SOMM allowed us to capture neuroactivity across 11 cortical regions, including the primary somatosensory cortex, primary visual cortex, and retrosplenial cortex during free behavior. In parallel we identified the associated neuroactivity with specific periods of social interactions by tracking animal behavior using DeepLabCut.
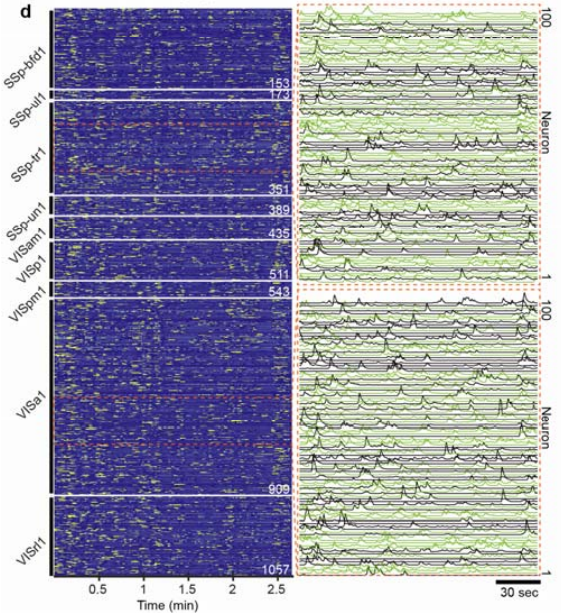
Figure 4 | Temporal activity rendering of 1057 neurons inferred by the processing pipeline in a 2.7-minute recording. Two zoom-in panels showed example traces (each with 100 896 neurons).
Video 1 | Overhead motion video of freely behaving mice engaged in social interactions. Motion tracing was performed with DeepLabCut, the SOMM was affixed to one animal with head bars while while interacting with cospecies, and the calcium traces were recorded simultaneously.
In summary SOMM is a compact and lightweight device designed on the basis of diffractive optics. SOMM allows to capture large-scale neuronal population activity at cellular resolution in mice during free-behavior and interactions with cospecies. This was achieved by SOMM’s design that relies on a DOE optimized to optimally navigate the optical design tradeoffs and meet the constraints of the desired form factor enabling single cell resolution imaging of neuroactivity of over 10,000 neurons at 4µm optical resolution within a FOV of 3.6 × 3.6 mm2 and 300 µm DOF at a weight of less than 2.5 grams
Relevant Publication:
Y. Zhang*, L. Yuan*, J. Wu, T. Nöbauer, R. Zhang, G. Xiao, M. Wang, H. Xie, Q. Dai, and A. Vaziri
A Systematically Optimized Miniaturized Mesoscope (SOMM) for large-scale calcium imaging in freely moving mice
Nature Biomedical Engineering (2024). doi: 10.1101/2024.02.19.581043
https://www.nature.com/articles/s41551-024-01226-2
