Optical trapping is a single molecule technique that allows to study mechanical properties of single polymer molecules, proteins, or individual chemical bonds. The technique belongs to the class of force spectroscopy tools which also commonly include Atomic Force Microscopy (AFM) and magnetic tweezers.
In a typical experiment controlled forces are applied to a single proteins complexes such as molecular motor and the dynamics of their behavior is studied. In practice it is difficult to apply force to a molecule directly, thus a handle is always used. We employ surface tether assays in which the force to a molecular motor is applied through two tethers, one that binds to a 0.5-micron size plastic bead and another that binds to a microscope coverslip (Fig. 1).
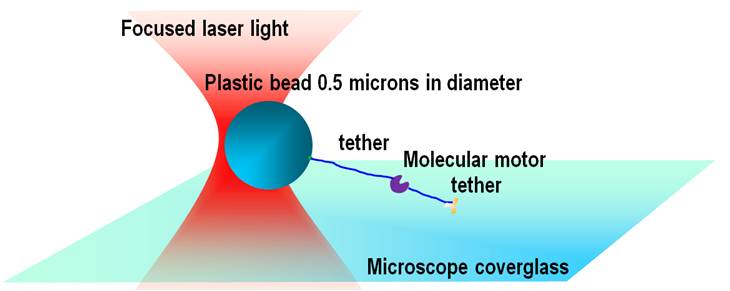
This arrangement allows applying forces in pico-newton range and reliably detect the displacement of the motor protein’s displacement as small as few angstroms with high temporal resolution. Special care needs to be taken of different sources of noise. We have built an optical trapping instrument that has long term drift less than 5 nm/min and allows detecting displacements as small as 3 nm (Fig. 2).
Applications of the Optical Tweezers:
Optical tweezers have revolutionized our understanding of how molecular motors function and allowed to ask question previously unanswerable by other methods. However, there is still a big gap between our ability to apply controlled forces to single molecules and our understanding of how this force affects states of the molecular motor and hence conclusions that we can draw about the molecular mechanism of its action.
We are currently working on two aspects; First, we are interested in designing new methods based on optical tweezers that should allow us to extract additional new information about molecular motors. Second, we are applying our tools for studying DNA polymerase and RNA polymerase molecular motors.
Relevant Publications:
C. Mieck, M. Molodtsov, K. Drzewicka, B. van der Vaart, G. Litos, G. Schmauss, A. Vaziri, and S. Westermann
Non-catalytic motor domains enable processive movement and functional diversification of the kinesin-14 Kar3
eLife, 10.7554/eLife.04489 (2015).
(Download)
McIntosh, J. Richard, Maxim I. Molodtsov, and Fazly I. Ataullakhanov
Biophysics of mitosis
Quarterly Reviews of Biophysics 45.2, 147-207 (2012).
(Download)
